We study structure and function of biological macromolecular systems. One of the current projects is the identification and energetic characterization of gas permeation pathways through membrane channels. We employ multi-scale molecular dynamics simulations, namely explicit/implicit/umbrella ligand sampling simulations, and in sillico mutagenesis to provide a complete map of gas migration pathways in membrane proteins. We also work on the computational modeling of membrane proteins to characterize the dynamic mechanisms of these proteins at atomic resolution.
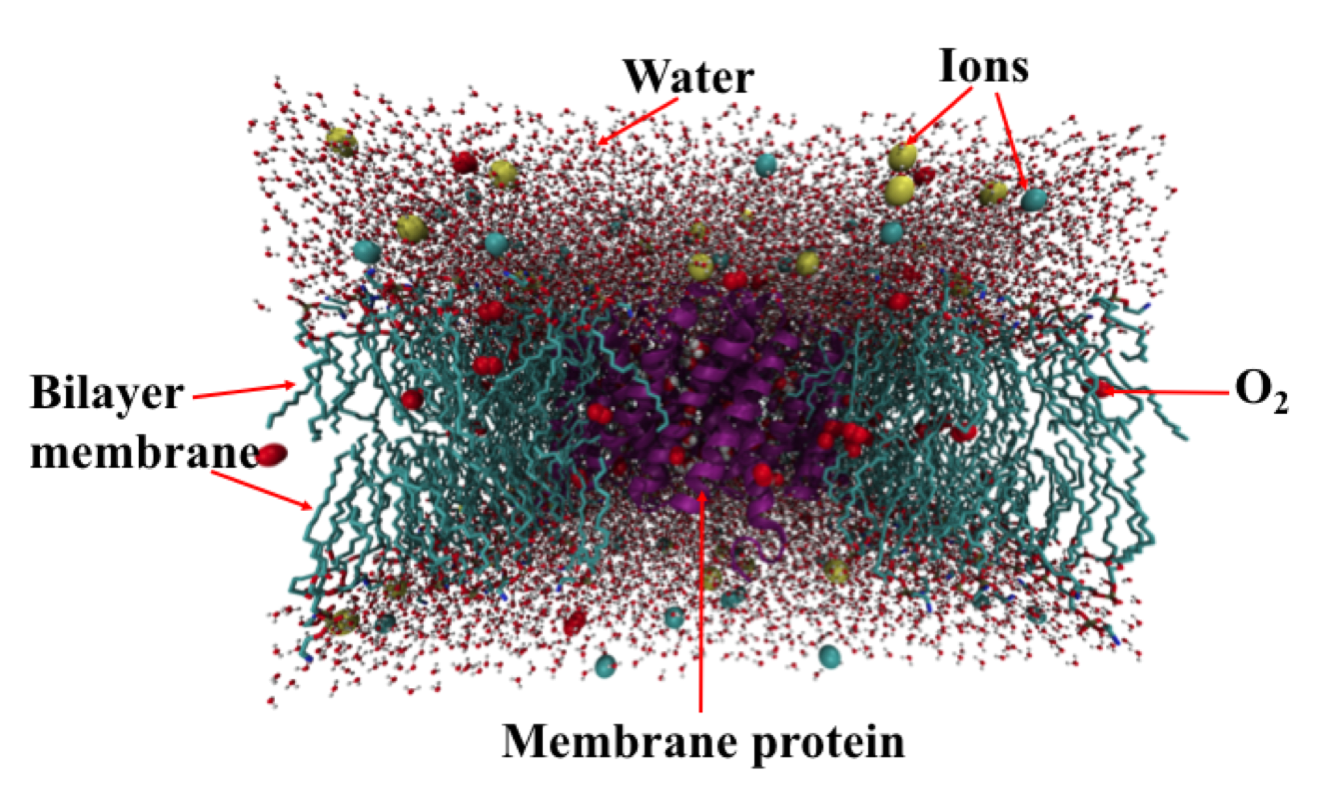 A typical molecular dynamic simulation system of a membrane protein. The periodic unit cell in this simulation includes an aquaporin protein, bilayer membrane, gas species, and water molecules.
A typical molecular dynamic simulation system of a membrane protein. The periodic unit cell in this simulation includes an aquaporin protein, bilayer membrane, gas species, and water molecules.
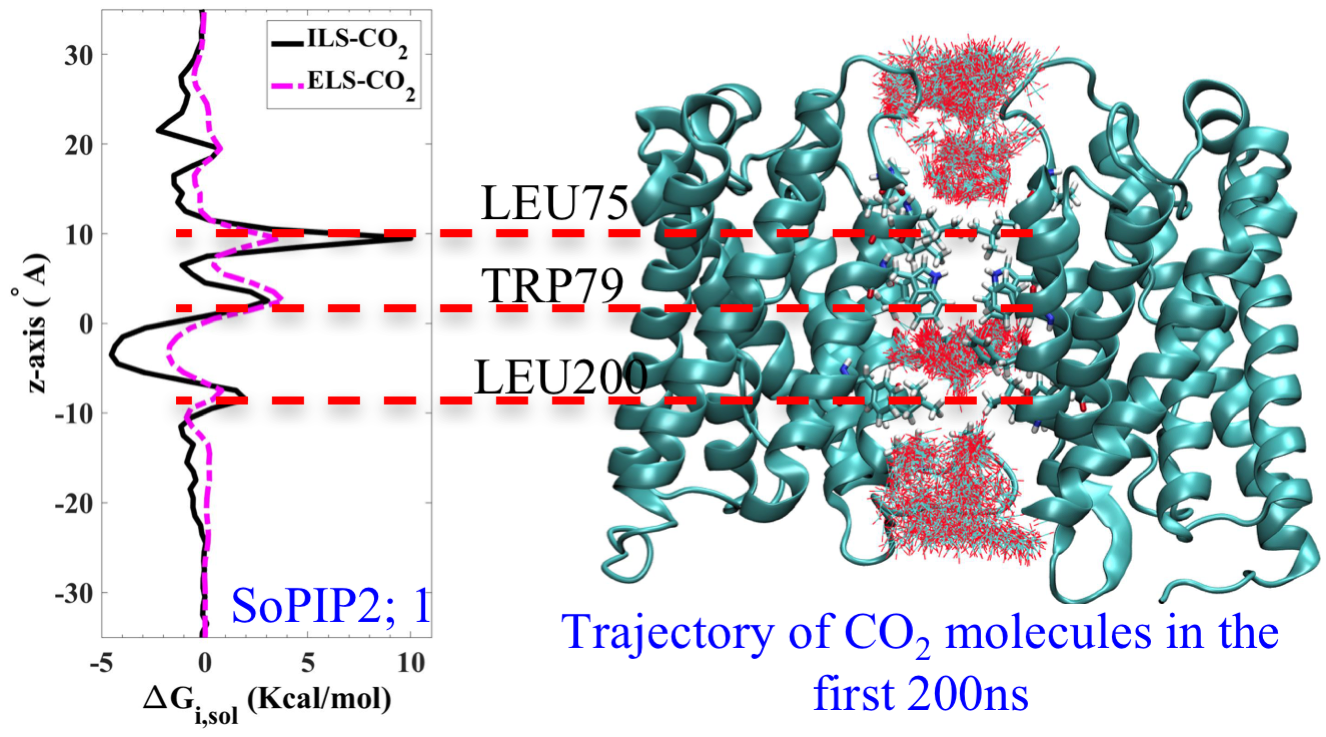 Energetic characterization of gases migration pathways for a membrane aquaporin using explicit and implicit sampling methods. This work was done in collaboration with Dr. Tajkhorshid’s lab.
Energetic characterization of gases migration pathways for a membrane aquaporin using explicit and implicit sampling methods. This work was done in collaboration with Dr. Tajkhorshid’s lab.
Related Publications
- Raeisi Najafi A., Mahinthichaichan P., Tajkhorshid E.; “Identification and Energetic Characterization of Gas Permeation Pathways through a Plant Water Channel” Biophysical Journal, Vol. 112, no. 3, pp. 423a, 2017.
